
Sporobolus stapfianus is a member of one of the largest and most important plant families, the Poaceae, but, alas, as Gaff et al. have put it, it is “obscure”.1 Despite not having a large devoted fan club, it is widely distributed in dry savannahs in central/eastern and southern Africa, Nigeria and Yemen at elevations from ca. 90 to 1700 m ASL.2 Most of the specific ecological information regarding it concerns its role as a forage species and as a component of the savannah and woodland communities, especially in Uganda.
In a survey of grazing by wild animals conducted Queen Elizabeth National Park (southwest Uganda) in the mid-1950s, for example, S. stapfianus was found throughout the park, from areas considered lightly grazed, to moderately overgrazed to seriously overgrazed, with highest representation in the moderately overgrazed zone. There, it represented 19% of the total range vegetation in the wet season, and over 45% at the end of the dry season.3 This may reflect its resilience, or it may reflect that it is not especially appreciated by herbivores. The latter is suggested by results of studies in northeast Uganda around cattle watering holes.4There, species composition and plant size were affected by distance from a watering hole itself. More desirable species increase in vitality with greater distance (indicating that grazing pressure is heavier close-in) while less desirable species decrease in vitality with distance, meaning, perhaps, that they can’t compete with other grasses in the absence of heavier grazing. Sporobolus stapfianus is in the latter group.
Overall, S. stapfianis may simply have too little biomass productivity to be of great importance as fodder for stock if there are other choices. However a recent study in the western United States concluded that its forage quality traits were sufficiently high that it could be considered as a potential forage in those semi-arid and arid regions where irrigation resources for the growth of alternative forage is limited.5
What really distinguishes S. stapfianus from other members of the genus, however, is its response to eXtreme drought: it is a resurrection plant. In this case, the desiccation tolerance is not apparent in all tissues, but is restricted to immature leaves of intact plants6, and an as yet uncertain portion of the root system. Young leaves can tolerate drying to at least -250MPa leaf water potential for at least a year, comparable to the dryness of seeds. In addition, S.stapfianus is reportedly tolerant of ionizing radiation, extreme temperatures and salinity to at least 215 mol m‑3 NaCl. In fact, it has been reported to survive air-dryness following 3 weeks pretreatment with salinities up to 200 mol m‑3.7
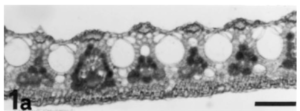

Overall, the cellular-level morphological alterations during dehydration in S. stapfianus are relatively small. Even in dehydrated leaves, the chloroplasts remain largely intact although the thylakoids become irregularly distributed and packed in large grana. The loss of chlorophyll itself is only partial, to about 40% of the fully hydrated leaf value. Within 48 h of rehydration, the chlorophyll content returns to 90% of the control leaf values and chloroplast organization is fully restored. Also during dehydration, the chloroplasts lose their starch, probably associated with the accumulation of sucrose as a vitrifying and protecting compound.8
Metabolomics of desiccation
At least two studies have examined changes in metabolites during dehydration2 and rehydration6 in S. stapfianus. Both of those are studies are extensive and impressive, and the summary here hardly does them justice. Go read them.
In the fully hydrated state, in comparison with a closely related and co-occurring sister species, S. pyramidalis, S. stapfianus had higher concentrations of osmolytes, lower concentrations of metabolites associated with energy metabolism, and higher concentrations of nitrogen metabolites. Because of the roles such compounds play in dehydrated tissue, this result suggests that S. stapfianus is primed metabolically for the stress. (Priming has been noted in a number of stress tolerant species for a number of stresses). With dehydration to a leaf RWC to 60%, S. stapfianus shifted toward the production of protective compounds (putative cellular osmolytes, including several amino acids, a number of sugars and sugar alcohols, and antioxidants). For the most part, concentrations of the same compounds in S. pyramidalis remained unchanged. At RWC lower than 40%, the S. stapfianus leaf metabolome was strongly directed toward antioxidant production, nitrogen remobilization, ammonia detoxification, and soluble sugar (sucrose) production.2 S. pyramidalis was, alas, no longer with us by that point.
Sucrose accumulation during dehydration would, on the face of it, be a potentially major energy source for the reactivation of metabolism during rehydration. During the rehydration process, energy must be provided for restoration of metabolic activity and repair of desiccation-induced damage, and respiration is rapidly reactivated following watering. The role of sucrose in this is unclear, however, because its breakdown is not apparent in the initial phase of rehydration.6 Later, however, when sucrose levels do fall, the concentrations of hexoses (glucose and fructose) remain unchanged, suggesting that at that point, sucrose is indeed a major energy source.
In the dehydrated state, S. stapfianus also seems to be primed to reactivate metabolism. For example, hexokinase activity at the start of rehydration was 4.5-fold higher than that in the fully hydrated controls, having been unregulated during dehydration. Phosphofructokinase (PFK) showed a similar pattern. Both activities remained high during 10-d of rehydration. Respiration also increased immediately on rewatering and before there was significant sucrose breakdown. The levels of malate dehydrogenase (MDH) and glutamate dehydrogenase (GDH) were high enough that the use of amino acids as respiratory substrates would apparently allow this.
A special note about proline – Reports of the accumulation of proline with water stress go back well into the 1960s, and despite the widespread tendency to assay this one amino acid in drought or salt experiments, its direct relationship with stress tolerance actually is more than a little questionable. Studies with S. stapfianus are a good lesson in this regard.
In this species, with desiccation, Tymms and Gaff reported in 1979 that proline increased 5.6 fold, intermediate for the group of 11 resurrection species they studied. With one exception, no species increased proline contents by more than 10%, and three either “broke even” or lost proline. These values can be compared with those of non-DT plants which accumulate up to 360 fold more proline with water stress. If one is looking for actual meaning in changes in amino acid levels, perhaps as significant was the relationship between proline increases and increases in total soluble non-protein N.9,10 In S. stapfianus, the increase in proline accounted for only 5% of the total non-protein N increase, i.e. its overall contribution to that pool changed little with desiccation. The authors concluded that the increased proline played no role in DT; rather, it reflected the controlled degradation of protein (autophagy) preparatory to desiccation.11
Sugars – In S. stapfianis, as in other resurrection plants, the accumulation of sugars, particularly sucrose, but also raffinose, stachyose and myo-inositol are adaptive as they are all are involved in vitrification of the cytosol and can served as energy reserves on rehydration.6 In this species, the sugars and sugar alcohols are the only case in which metabolite and transcript levels correlate well during desiccation and rehydration.12 In roots, sugar alcohols also increase (especially galactinol) as does trehalose, perhaps as a signaling molecule as it has no significance as an osmoticum. That it is carbohydrate accumulation that matters and not necessarily any single sugar or alcohol is implied by the fact that in some species, e.g. Selaginella, mannitol and galactinol increase with desiccation, but not the sugars.13
Transcriptomics and Genomics
Unlike other species dealt with on this site until now, S. stapfianus has a much larger genome (haploid=1.3Gb) and it is tetraploid rather than diploid. As a result of this additional complexity, there is as yet no sequencing project or genome assembly for the species.
Excluding chloroplast genes (e.g. rbcL), internal transcribed spacers and patents, GenBank currently contains only 25 nucleotide sequences for S. stapfianus. All are for genes whose expression is markedly affected by desiccation, and all are from Le et al.14 or Neale et al.15Following that link will give the latest results whenever more are added. In addition, there are two BioProjects from a transcriptome shotgun assembly using 454 sequencing. Those results have been published.12 Following that link will also give any additional projects as they appear.
This page was put composed by John Cheeseman who is grateful for the correspondence, discussions and assistance of Mel Oliver, Don Gaff and Concetta Vazanna. Additions, corrections and suggestions are welcome.
Sporobolus stapfianus appears as the major focus of only a few articles indexed in PubMed although it is much more broadly represented in articles dealing with DT in general. The following is a partial list of publications from that collection, focussed on studies emphasizing a molecular aspect. The list was generated by a search of PubMed; additional publications not indexed in PubMed are, of course, not included. Abstracts and/or full text versions can be accessed by clicking doi, PMID or PMC numbers. For a list of all PubMedCentral (PMC) listed publications including S. stapfianus, click here.
- Chávez Montes, RA, Haber, A, Pardo, J, Powell, RF, Divisetty, UK, Silva, AT et al. (2022) A comparative genomics examination of desiccation tolerance and sensitivity in two sister grass species. Proc Natl Acad Sci U S A 119:. doi: 10.1073/pnas.2118886119. PubMed PMID:35082155 PubMed Central PMC8812550.
- Yobi, A, Batushansky, A, Oliver, MJ, Angelovici, R (2019) Adaptive responses of amino acid metabolism to the combination of desiccation and low nitrogen availability in Sporobolus stapfianus. Planta 249:1535-1549. doi: 10.1007/s00425-019-03105-6. PubMed PMID:30725176 .
- Yobi, A, Schlauch, KA, Tillett, RL, Yim, WC, Espinoza, C, Wone, BW et al. (2017) Sporobolus stapfianus: Insights into desiccation tolerance in the resurrection grasses from linking transcriptomics to metabolomics. BMC Plant Biol 17:67. doi: 10.1186/s12870-017-1013-7. PubMed PMID:28351347 PubMed Central PMC5371216.
- Griffiths, CA, Gaff, DF, Neale, AD (2014) Drying without senescence in resurrection plants. Front Plant Sci 5:36. doi: 10.3389/fpls.2014.00036. PubMed PMID:24575108 PubMed Central PMC3922084.
- Islam, S, Griffiths, CA, Blomstedt, CK, Le, TN, Gaff, DF, Hamill, JD et al. (2013) Increased biomass, seed yield and stress tolerance is conferred in Arabidopsis by a novel enzyme from the resurrection grass Sporobolus stapfianus that glycosylates the strigolactone analogue GR24. PLoS One 8:e80035. doi: 10.1371/journal.pone.0080035. PubMed PMID:24224034 PubMed Central PMC3818285.
- Oliver, MJ, Guo, L, Alexander, DC, Ryals, JA, Wone, BW, Cushman, JC et al. (2011) A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus. Plant Cell 23:1231-48. doi: 10.1105/tpc.110.082800. PubMed PMID:21467579 PubMed Central PMC3101564.
- Oliver, MJ, Jain, R, Balbuena, TS, Agrawal, G, Gasulla, F, Thelen, JJ et al. (2011) Proteome analysis of leaves of the desiccation-tolerant grass, Sporobolus stapfianus, in response to dehydration. Phytochemistry 72:1273-84. doi: 10.1016/j.phytochem.2010.10.020. PubMed PMID:21109273 .
- Gaff, DF, Blomstedt, CK, Neale, AD, Le, TN, Hamill, JD, Ghasempour, HR et al. (2009) Sporobolus stapfianus, a model desiccation-tolerant grass. Funct Plant Biol 36:589-599. doi: 10.1071/FP08166. PubMed PMID:32688672 .
- Li, FH, Fu, FL, Sha, LN, Li, WC (2007) Identification of drought-responsive genes from maize inbred lines. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 33:607-11. . PubMed PMID:18349516 .
- Whittaker, A, Martinelli, T, Farrant, JM, Bochicchio, A, Vazzana, C (2007) Sucrose phosphate synthase activity and the co-ordination of carbon partitioning during sucrose and amino acid accumulation in desiccation-tolerant leaf material of the C4 resurrection plant Sporobolus stapfianus during dehydration. J Exp Bot 58:3775-87. doi: 10.1093/jxb/erm228. PubMed PMID:18057046 .
- Martinelli, T, Whittaker, A, Masclaux-Daubresse, C, Farrant, JM, Brilli, F, Loreto, F et al. (2007) Evidence for the presence of photorespiration in desiccation-sensitive leaves of the C4 'resurrection' plant Sporobolus stapfianus during dehydration stress. J Exp Bot 58:3929-39. doi: 10.1093/jxb/erm247. PubMed PMID:18037680 .
- Martinelli, T, Whittaker, A, Bochicchio, A, Vazzana, C, Suzuki, A, Masclaux-Daubresse, C et al. (2007) Amino acid pattern and glutamate metabolism during dehydration stress in the 'resurrection' plant Sporobolus stapfianus: a comparison between desiccation-sensitive and desiccation-tolerant leaves. J Exp Bot 58:3037-46. doi: 10.1093/jxb/erm161. PubMed PMID:17901195 .
- Martinelli, T (2008) In situ localization of glucose and sucrose in dehydrating leaves of Sporobolus stapfianus. J Plant Physiol 165:580-7. doi: 10.1016/j.jplph.2007.01.019. PubMed PMID:17765358 .
- Le, TN, Blomstedt, CK, Kuang, J, Tenlen, J, Gaff, DF, Hamill, JD et al. (2007) Desiccation-tolerance specific gene expression in leaf tissue of the resurrection plant Sporobolus stapfianus. Funct Plant Biol 34:589-600. doi: 10.1071/FP06231. PubMed PMID:32689387 .
- Whittaker, A, Bochicchio, A, Vazzana, C, Lindsey, G, Farrant, J (2001) Changes in leaf hexokinase activity and metabolite levels in response to drying in the desiccation-tolerant species Sporobolus stapfianus and Xerophyta viscosa. J Exp Bot 52:961-9. doi: 10.1093/jexbot/52.358.961. PubMed PMID:11432913 .
- O'Mahony, PJ, Oliver, MJ (1999) The involvement of ubiquitin in vegetative desiccation tolerance. Plant Mol Biol 41:657-67. doi: 10.1023/a:1006330623364. PubMed PMID:10645725 .
- O'Mahony, PJ, Oliver, MJ (1999) Characterization of a desiccation-responsive small GTP-binding protein (Rab2) from the desiccation-tolerant grass Sporobolus stapfianus. Plant Mol Biol 39:809-21. doi: 10.1023/a:1006183431854. PubMed PMID:10350094 .
- Clugston, SL, Daub, E, Honek, JF (1998) Identification of glyoxalase I sequences in Brassica oleracea and Sporobolus stapfianus: evidence for gene duplication events. J Mol Evol 47:230-4. doi: 10.1007/pl00006380. PubMed PMID:9694672 .
- Wood, JN, Gaff, DF (1989) Salinity studies with drought-resistant species of Sporobolus. Oecologia 78:559-564. doi: 10.1007/BF00378748. PubMed PMID:28312187 .
